Protein Engineering

- account_circleSohei ItoPhD, Assoc. Prof.
- account_circleDaisuke FujinamiPhD, Research Asst. Prof.
- Website:https://www.tanpaku-lab.com/
- Mail:itosohei@u-shizuoka-ken.ac.jp
- Phone:+81-54-264-5576
Investigation of the structure and function of enzymes
Application of protein engineering techniques to the food sciences
- 1. Studies on the structure of proteins used in food engineering
X-ray crystallography is our most powerful tool for showing the three-dimensional structure of a protein. We analyzed the structures of many proteins linked to human disease.
- 2. Protein engineering of enzymes relating to plant secondary metabolism
Plants produce many substances to attract pollinators and seed disseminators and to repel parasites, predators, and herbivores. Terpene synthase is one of the most important enzymes concerning biosynthesis of plant volatiles. We have successfully isolated and characterized from tea and citrus plants several genes related to terpene synthases.
- 3. Protein engineering of amylases and their related enzymes
Sugar-related enzymes such as amylases and glucosidases are employed to produce sweetener from starch. The enzymes used for this purpose should have high thermostability. We are researching suitable enzymes from thermophilic bacteria.
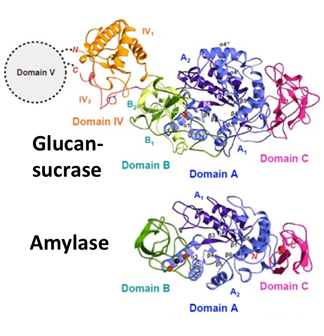
- Figure 1
- Crystal structure of Glucansucrase and amylase. Ribbon diagram of Glucansucrase from Streptococcus mutans (upper left). Ribbon diagram of α-amylase from Bacillus licheniformis (left below). Substrate-binding site with acarbose (right). Domain IV, A and B are composed of chains IV1 (orange) and IV2 (salmon), chains A1 (blue) and A2 (purple) and chains B1 (green) and B2 (yellow green), respectively. Acarbose is a pseudotetrasaccharide and a strong inhibitor for Glucansucrases. The glycosyl intermediate in the structure of another amylase family member is shown in gray.
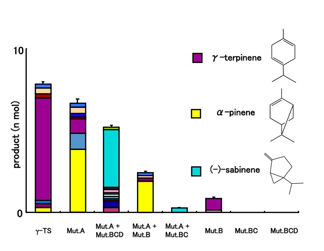
- Figure 2
- Rational conversion of product specificity in a Citrus unshiu monoterpene synthase. Introduction of mutations near the active site results in changes in the products formed. Some mutated enzymes produced α-pinene and sabinene in lieu of γ-terpinene.
References
- Biosci. Biotechnol. Biochem. 75, 1245-1248 (2011)
- J. Mol. Biol. 408, 177-186 (2011)
- J. Am. Chem. Soc. 132, 824-832 (2010)