Food Bioinformatics

Biotechnology and Bioinformatics
Development of new technology to design highly functional biomaterials
- 1. Development of novel sequence design method to generate highly functional proteins.
We are now developing new computational tools to design highly functional biomaterials by analyzing a huge amount of DNA or protein sequence data registered in public database. Many of highly functional proteins could be generated by the collaboration researches.
- 2. Establishment of enzymatic synthetic approach of fine chemicals
We try to screen new enzymes metabolizing L-amino acids from sequence database utilizing the developed tools. The screened enzymes are applied to synthesize fine chemicals (such as D-amino acid derivatives) of which precursors are L-amino acids.
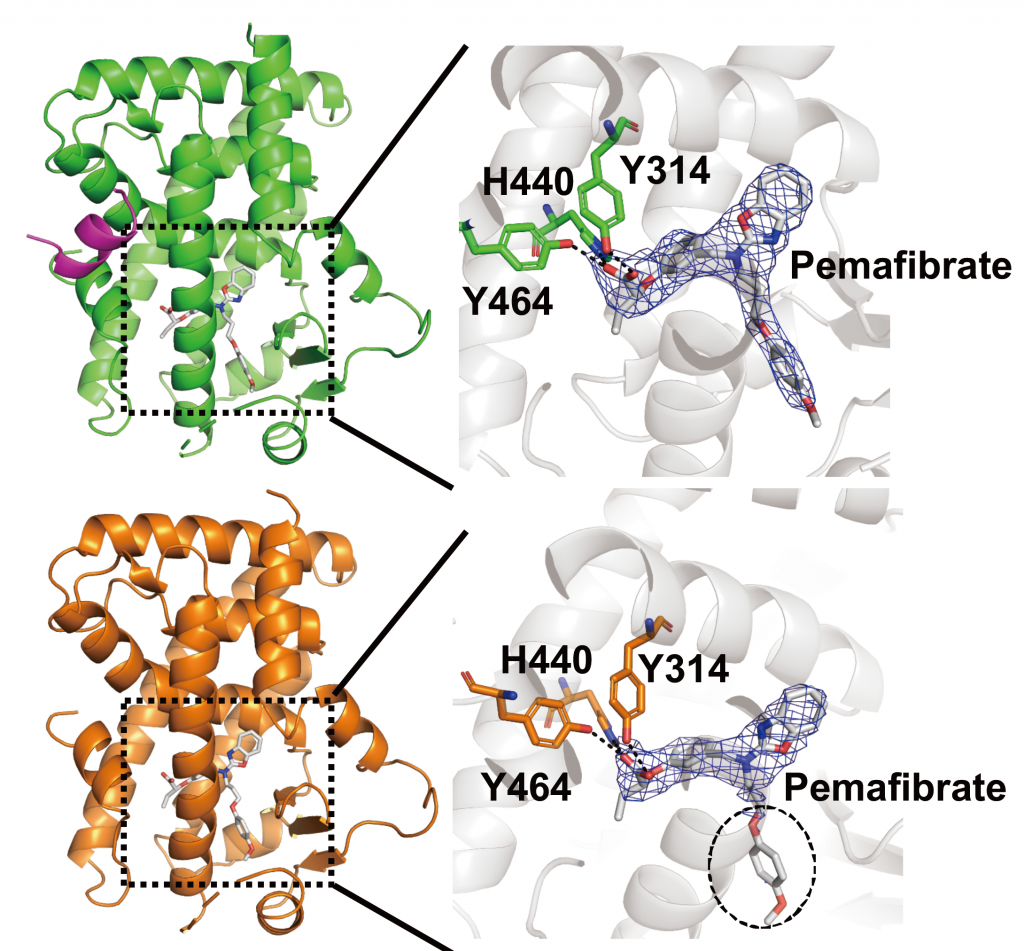
- 3. Structural and functional analysis of nuclear receptors
Structural and functional analysis for several of nuclear receptors (such as RXRα and PPARs) are performed by combinational analysis of X-ray crystallography, computational chemical and biochemical approaches.
- Figure 1
- Structure and functional analysis of new L-amino acid oxidase (ArtLAAO) assigned by in silico enzyme screening. ArtLAAO can apply to synthesize enantio-pure D-amino acid derivatives from L- or racemic amino acid derivatives (ACS Catal., 2019, Commun. Chem., 2020).

- Figure 2
- Molecular mechanism of selective PPARα modulator, pemafibrate. We can elucidate molecular mechanism why pemafibrate can form stable interaction with ligand binding domain of PPARα (IJMS, 2020).
References
- Commun. Chem. 3(1), 181, (2020)
- Biochemistry, 59(40), 3823-3833, (2020)
- IJMS, 21(1), 361, (2020)
- ACS Catalysis, 9(11), 10152-10158, (2019)
- J. Chem. Inf. Model., 59(1), 25-30, (2019)