Physiology

- account_circleHisayoshi HayashiPhD, Asst. Prof.
- account_circleHEMPSTOCK WendyPhD, Research Asst. Prof.
- Website:https://dfns.u-shizuoka-ken.ac.jp/labs/physiol/
- Mail:hayashih@u-shizuoka-ken.ac.jp
- Phone:+81-54-264-5532
Health from a healthy gut
Healthy living requires an understanding of intestinal function
- 1. Elucidation of the physiological significance of the upper small intestine energy-sensing system.
While it is generally accepted that the small intestine possesses nutrient absorption functions throughout its length, experimental evidence to support this notion has not been presented. Unlike the lower small intestine, the nutrient absorption mechanism of the upper small intestine is only active during fasting and not during feeding. It is thought that the upper small intestine senses the energy status of the body and acts as needed. This study aims to elucidate the physiological significance of the energy-sensing switch mechanism in the upper small intestine and its molecular mechanism.
- 2. Investigation of the digestive tract barrier mechanism.
The digestive tract epithelium selectively absorbs necessary nutrients while also providing a barrier function against foreign substances and invading bacteria from the external environment. The maintenance of this barrier function is dependent on tight junctions between cells. The major structural proteins in the tight junctions between two cells are members of the claudin family, which form a zipper-like structure that seals the intercellular space. In contrast, a different structure has been observed in the three-cell junctions where three epithelial cells meet. Recently, tricellulin and angulin family proteins have been identified as the major structural proteins in three-cell tight junctions. Therefore, the roles of each protein in barrier function and nutrient absorption are being investigated using mice lacking angulin or claudin.
- 3. Establishment of a monolayer epithelial function measurement system derived from small intestinal organoids
Caco-2 cells, a model cell line of the small intestine, and samples extracted from animals have been used to evaluate the absorption and secretion functions of the small intestine in vitro. However, Caco-2 cells are derived from the large intestine, and it is difficult to quantitatively measure absorption function alone due to the three-dimensional structure of the villi in the extracted small intestine. Recently, the culture technique of "small intestinal organoids," which use isolated intestinal stem cells, has been developed to mimic the intestinal tissue in vivo. Small intestinal organoids are spherical structures with the lumen facing inward, and it is difficult to administer the nutrients to be measured. Therefore, we aim to establish a monolayer culture method for small intestinal organoids and develop an evaluation system for nutrient absorption function.
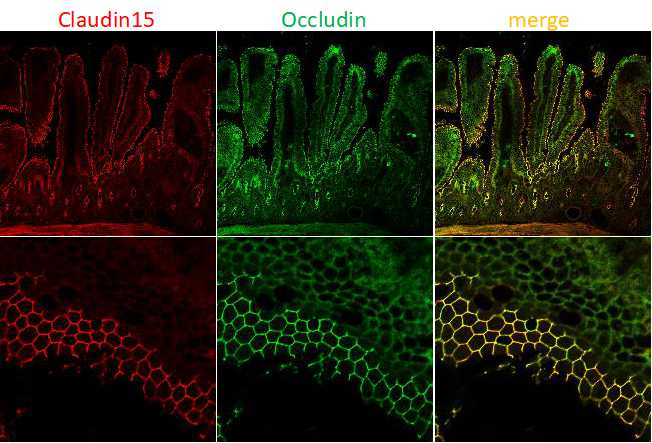
- Figure 1
- Immunofluorescence image of claudin-15 (red) and occludin (green) in the murine small intestine. Claudins form paracellular barriers and determine the properties of intestinal epithelia.
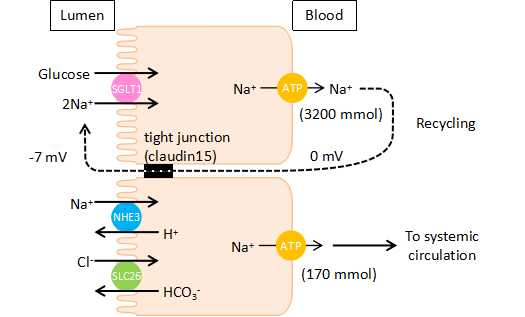
- Figure 2
- Schematic illustration of Na+ recycling mechanisms in the murine small intestine. Na+, which is absorbed by Na+-dependent glucose cotransport, is recycled back into the lumen via paracellular Na+ pathways.
References
- Am. J. Physiol., 324, R645-655 (2023)
- Cell Struct Funct.,48,1-17 (2023)
- J. Vis. Exp.,171.,Doi: 10.3791/62468 (2021)
- Sci. Rep., 10, 10374 (2020)
- Int. J. Mol. Sci., 21, 376 (2020)
- Am. J. Physiol., 315, G799-809 (2018)
- Sci. Rep., 7, 12223 (2017)
- PLOS ONE, 8(2), e55623 (2013)