Microbiology
-1-120x136.jpeg)
Development of immune suppression / immune tolerance by microorganisms
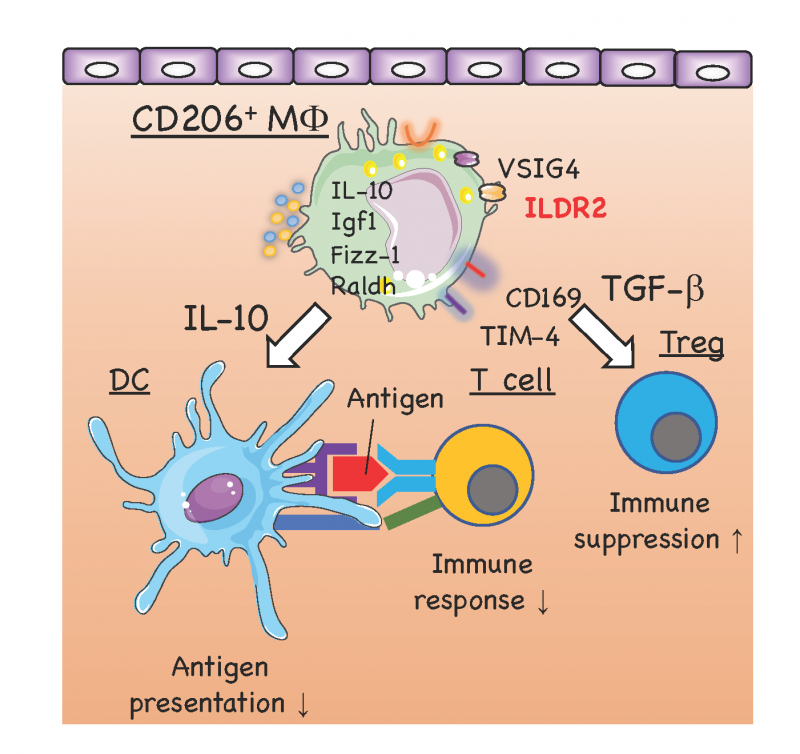
- 1. Analyses of immune suppressive sublingual macrophages
With repeated antigen application to the sublingual mucosa, we found that immunosuppressive macrophages increase in the sublingual mucosa and expression of unique immune checkpoint molecules that characterize these cells. We will analyze the dynamics of these macrophages and the expression of the immune checkpoint molecule by applying microorganisms.
- 2. Analyses of differentiation of type 1 regulatory T cells
Among helper T cell subsets, we found that the differentiation of type 1 regulatory T cell (Tr1) cells, which produce the immunosuppressive cytokine IL-10, are regulated by phosphoinositide 3-kinase (PI3K) pathway. We will look for microorganisms that act on this pathway and try to regulate Tr1 cell differentiation.
- 3. Analyses of the mechanisms inducing immune paralysis
After sepsis caused by infection, the systemic immunity is suppressed (called immune paralysis), which is further aggravated by a secondary infection. We try to clarify the dynamics of the cells involved in this immunosuppression and to find a way to break the immune paralysis.
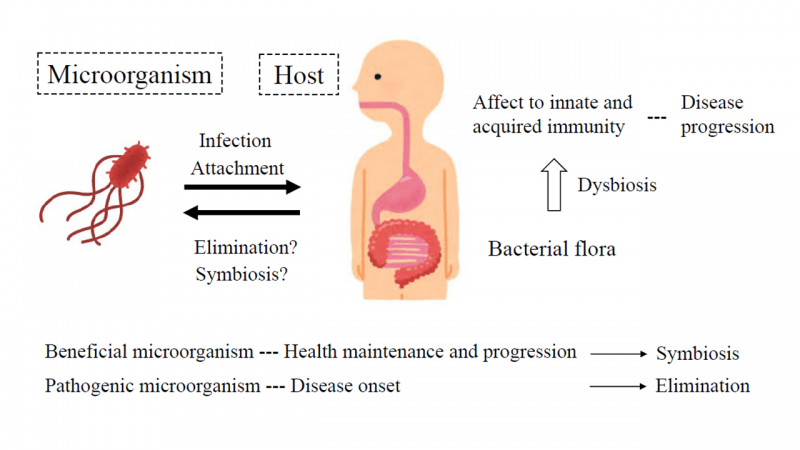
- Figure 1
- Host-microorganism interaction to maintain the immune homeostasis
- Figure 2
- Tolerogenic macrophages appeared in the oral mucosa after repeated antigen application to the sublingual mucosa
References
- Biochem. Biophys. Res. Commun., 742, 151009 (2025)
- Infect. Immun., 89, e00771-20 (2021)
- Int. Immunol., 32, 509-518 (2020)
- Immunology, 480, 114-119 (2017)